RNA-Seq, a path to biological insight
RNA-Seq has become the method of choice for exploring a target organism's gene expression.
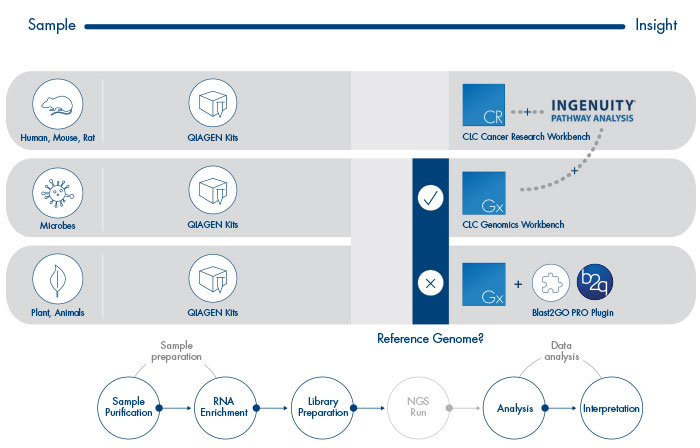
In a recent survey the majority of scientists using Next-generation Sequencing (NGS) have emphasized the importance of RNA-Seq solutions for their work. QIAGEN is dedicated to guiding scientists from Sample to Insight with solutions that start with the processing of biological samples and end with the interpretation of RNA-Seq data.
Benefits of QIAGEN Bioinformatics
Data Analysis
CLC Workbenches allow life scientists, clinicians and bioinformaticians alike to carry out steps crucial for the analysis of RNA-Seq data, easily and compute resource-efficiently:
- Quality control of data
- Demultiplexing
- Quantitative mapping of RNA-Seq reads
- Normalization and statistical analysis of differential gene expression
RNA-Seq in organisms without reference genome becomes possible through the following additional features:
- De novo assembly of transcriptome
- Functional annotation of unknown transcripts via the Biobam Blast2GO Pro plugin
Interpretation
Results like statistical information, read mappings, and differential gene expression are visualized and can be browsed in the context of other genomic information, such as annotations, variants, or epigenetic information like ChIP-Seq results.
CLC Genomics Workbench and CLC Cancer Research Workbench seamlessly integrate with Ingenuity Pathway Analysis (IPA), for biological interpretation of gene expression results and complex omics data from human, mouse, or rat.
Ease of Use
Both, CLC Genomics Workbench and CLC Cancer Research Workbench offer an intuitive user experience and require no background in bioinformatics.
Join the webinar


Take a test drive
Scientific Spotlight
Try out our trusted solution for RNA-Seq Analysis in a fully functional trial accompanied by a set of tutorials offering step by step instructions for the analysis of your data. Analyze your own data or the example data set provided within the tutorial.
RNA-Seq Analysis Tutorials:
Part I Getting Started
Part II Non-Specific Matches, Mapping Modes and Expression measures
Part III Identifying differentially expressed genes and transcripts
Comprehensive transcriptome analysis of early male and female Bactrocera jarvisi embryos
Jennifer L. Morrow, et al. use CLC Genomics Workbench to analyze the transcriptome of the fruit fly Bactrocera jarvisi, a horticultural pest, generating new insights into candidate genes for transgenic pest-management strategies.
RNA-Seq analysis using de novo assembled transcriptome as reference
RNA Sequence Reveals Mouse Retinal Transcriptome Changes Early after Axonal Injury
Masayuki Yasuda, Yuji Tanaka, Morin Ryu, Satoru Tsuda, Toru Nakazawa
PLoS ONE 9(3): e93258. doi:10.1371/journal.pone.0093258
QIAGEN products
Sample Purification
RNA Enrichment
Library Preparation
Analysis
Interpretation

For up-to-date information and product-specific disclaimers, see the respective QIAGEN kit handbooks or user manual. QIAGEN kit handbooks and user manuals are avaliable at www.qiagen.com or can be requested from QIAGEN Technical Services or your locale distributer.